AP® Biology Unit 6 Review and Practice Test
Struggling with AP® Biology Unit 6 prep? We have got you covered. UWorld provides a thorough coverage of Gene Expression and Regulation, where our AP experts break down concepts like DNA, RNA, protein synthesis, genotype and phenotype, cell specialization, and more. So, whether you need help with AP Bio Unit 6 FRQs and MCQs, or simply want to understand concepts in detail, choosing UWorld can lead you to a 5.
Boost Your Confidence and Score High with Our AP Biology Unit 6 Review
Ready to achieve top scores with our AP® Biology Unit 6 review? Our coursework makes even the most challenging topics easier to understand. Our study guide, question bank, video lessons, and study tools are designed by experts to provide an all-round experience, helping you grasp concepts better and retain them longer.
Engaging Video Lessons
Better visuals often mean better learning and retention. UWorld provides engaging, colorful, and image-rich video lessons to help you learn faster, understand better, and avoid the confusion of text-heavy explanations.
Interactive Study Guides
Our Unit 6 AP Bio study guide is consistently aligned with the College Board® CED. Each topic comes with in-depth explanations, well-labelled diagrams, and check points to ensure you understand what you learn.
Try These AP Biology Unit 6 Practice Test Questions
Question
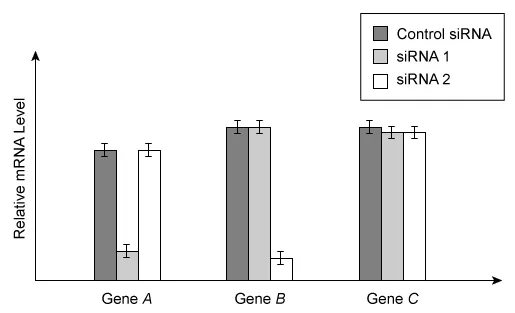
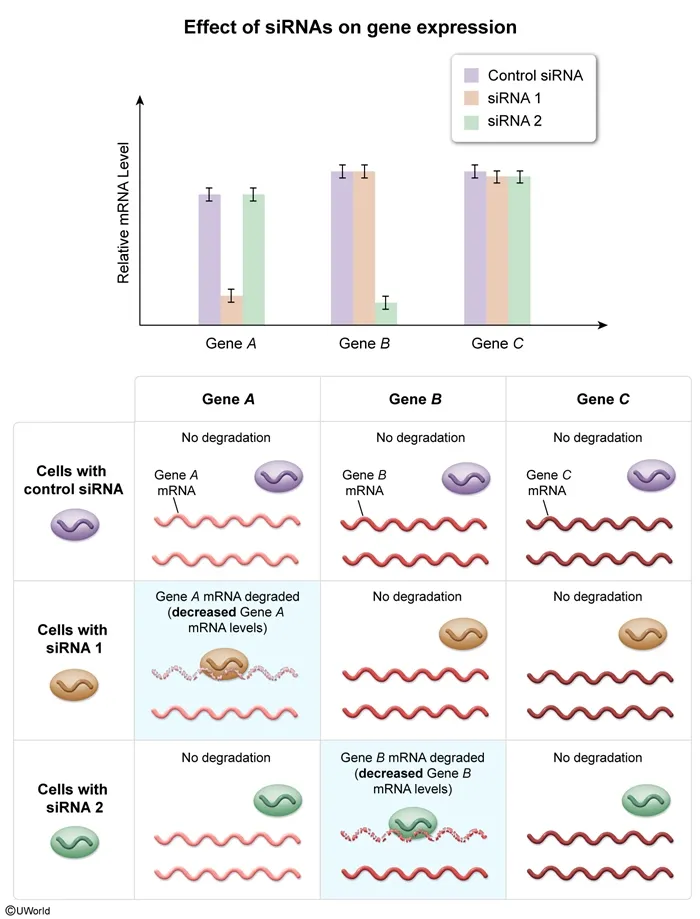
Some small interfering RNAs (siRNAs) are known to regulate gene expression in eukaryotic cells. In an experiment, two groups of eukaryotic cells were each stimulated to take up different siRNA molecules (siRNA 1 or siRNA 2). Both of these siRNAs are known to affect gene expression. A third group of cells was stimulated to take up control siRNAs, which were engineered to have no effect on gene expression. After 72 hours, the mRNA levels for three genes were measured in each group. The graph below shows the results of this experiment.
Which of the following statements about these siRNA molecules is best supported by the results presented in the graph?
| A. siRNA 1 binds and causes the degradation of Gene B mRNA; siRNA 2 binds and causes the degradation of Gene A mRNA. | |
| B. siRNA 1 binds and causes the degradation of Gene A mRNA; siRNA 2 binds and causes the degradation of Gene B mRNA. | |
| C. Both siRNA 1 and siRNA 2 bind the mRNA molecules produced from all three genes. | |
| D. Both siRNA1 and siRNA 2 have identical nucleotide sequences. |
Explanation
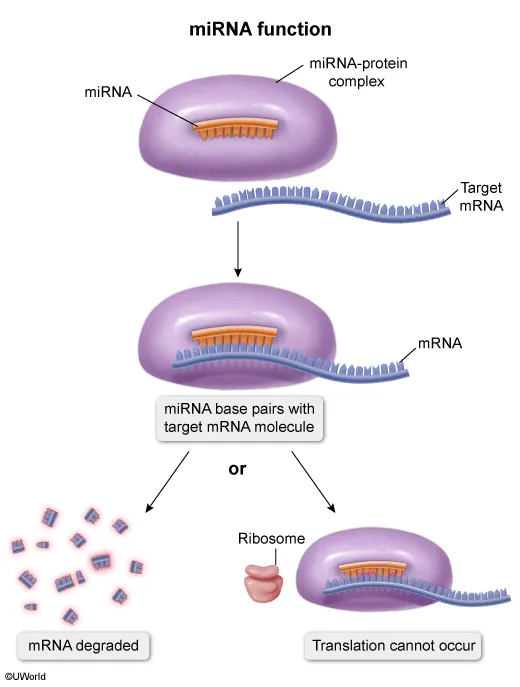
When genes are transcribed into mRNA transcripts, regulation of gene expression occurs post-transcriptionally (ie, at the level of the transcript). MicroRNAs (miRNAs) and small interfering RNAs (siRNAs) are small, noncoding elements that perform post-transcriptional regulation. These small RNAs often form complexes with specific proteins and bind complementary nucleotide sequences on target mRNA molecules, causing mRNA degradation or inhibition of translation.
In this question, two groups of eukaryotic cells were each stimulated to take up different siRNA molecules that affected gene expression. A third group of cells took up control siRNA that did not affect expression. The graph shows that, compared to cells with control siRNAs, cells that took up siRNA 1 exhibited decreased Gene A mRNA levels, whereas cells that took up siRNA 2 exhibited decreased Gene B mRNA levels.
Therefore, siRNA 1 binds and degrades Gene A mRNA, whereas siRNA 2 binds and degrades Gene B mRNA (Choice A).
(Choice C) siRNA 1 only decreased Gene A mRNA levels and siRNA 2 only decreased Gene B mRNA levels. In addition, neither siRNA 1 nor siRNA 2 affected Gene C expression compared to control cells. Therefore, siRNA 1 and siRNA 2 do not bind mRNA produced from all three genes to regulate their expression.
(Choice D) siRNA 1 and siRNA 2 decrease mRNA expression for different genes ( Genes A and B ), indicating that the nucleotide sequence complementary to the target mRNA is different (not identical) in each siRNA molecule.
Things to remember:
Small interfering RNAs (siRNAs) are small, noncoding RNA molecules that bind target mRNA molecules, often causing mRNA degradation.
Question
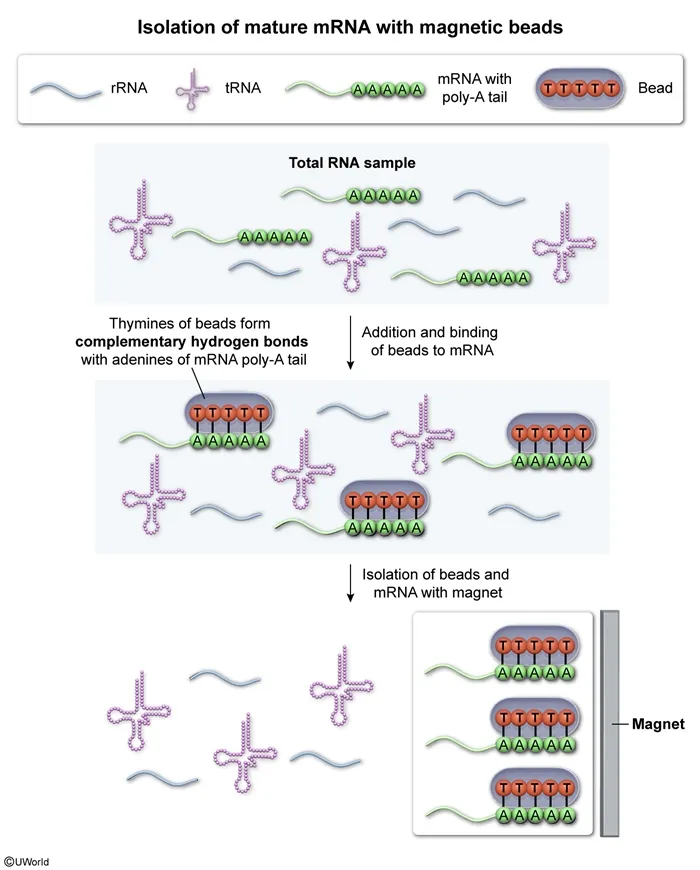
In a gene expression study, scientists extracted RNA samples from eukaryotic cells. These initial samples contained all types of RNA found within the cells. They then added a specialized solution containing microscopic magnetic beads to the RNA samples. These beads consisted of synthetic pieces of single-stranded DNA made up of repeated thymine nucleotides. These thymine nucleotides are able to form hydrogen bonds with mature mRNA molecules that have been modified in the nucleus. Once bound to mRNA, the beads were collected using a specialized magnet, allowing mature mRNA to be isolated from the initial RNA samples. Which of the following best describes how these beads most likely bound the mRNA molecules?
| A. The thymine nucleotides of the beads formed hydrogen bonds with the nucleotides in the poly-A tails of the mRNA molecules. | |
| B. The thymine nucleotides of the beads formed hydrogen bonds with many different types of nucleotides in the coding regions of the mRNA molecules. | |
| C. The thymine nucleotides of the beads formed hydrogen bonds with all three nucleotides of the universal start codons (AUG) in the mRNA molecules. | |
| D. The thymine nucleotides of the beads formed hydrogen bonds with the GTP caps of the mRNA molecules. |
Explanation
During eukaryotic gene expression, genes are initially transcribed into pre-messenger RNA (pre-mRNA) molecules in the nucleus. Prior to translation, pre-mRNA undergoes a series of modifications to form mature mRNA, including the splicing (ie, removal) of noncoding introns as well as the addition of a 3' poly-adenine (poly-A) tail and a 5' GTP cap.
Specifically, both the poly-A tail and GTP cap allow mature mRNA to exit the nucleus and enter the cytoplasm for translation. Once in the cytoplasm, the poly-A tail and GTP cap help prevent mRNA degradation and allow ribosomes to recognize and bind mRNA.
In this question, magnetic beads were added to eukaryotic RNA samples. These beads contained single-stranded DNA pieces made up of repeated thymine (T) nucleotides that bound mature mRNAs. Because all mature mRNAs have poly-A tails, the thymine nucleotides of the beads likely formed complementary hydrogen bonds with the adenine nucleotides of the mRNA poly-A tails.
(Choice B) In base pairing, thymine always pairs with adenine. Therefore, thymine nucleotides would not form hydrogen bonds with many different nucleotide types in mRNA coding regions.
(Choice C) All mRNA molecules have a universal start codon (AUG) where translation begins. Although one thymine nucleotide could bond with the adenine (A) in the start codon, thymine would not be complementary to uracil (U) or guanine (G).
(Choice D) Although all mature mRNAs have GTP caps, this region does not contain nucleotides complementary to thymine.
Things to remember:
In eukaryotic cells, pre-mRNA transcribed in the nucleus is modified to produce mature mRNA. Modifications include splicing (removing) introns and adding a 3' poly-A tail and 5' GTP cap.
Question
miR-1 is a small, noncoding nucleic acid that is single-stranded in nature and has ribose-containing nucleotides. miR-1 has been found in animal heart muscle cells (cardiomyocytes), which contract to facilitate the pumping action necessary to circulate blood throughout the body. Scientific research has proven that miR-1 posttranscriptionally regulates the production of HSP60, a protein known to prevent damage to other proteins in high-heat environments. Which of the following would best describe how miR-1 acts to regulate HSP60 expression in cardiomyocytes?
| A. miR-1 is read by ribosomes to generate HSP60 proteins. | |
| B. miR-1 pairs with proteins to form the ribosomes that will translate HSP60 proteins. | |
| C. miR-1 binds to complementary sequences on HSP60 mRNA, inhibiting its translation into proteins. | |
| D. miR-1 transfers incorrect codons to the ribosome, resulting in permanently shortened HSP60 proteins. |
Explanation
When genes are transcribed into messenger RNA (mRNA) transcripts, gene expression can be regulated at the level of the transcript. This kind of posttranscriptional regulation can be carried out by many elements, including small, noncoding RNA molecules such as microRNAs (miRNAs) and small interfering RNAs (siRNAs).
miRNAs bind to complementary sequences on target mRNAs to inhibit target gene expression. miRNA exerts its gene-silencing effects by forming a complex with a specific protein. The miRNA component of the protein-miRNA complex then binds to the complementary region of the target mRNA to either degrade the target mRNA or to block ribosome access to the target mRNA (prevents translation). In this way, miRNAs control which genes are expressed and which genes are not.
In this question, miR-1 most likely binds to complementary sequences on HSP60 mRNA, thereby inhibiting its translation into protein. Therefore, miR-1 posttranscriptionally regulates the expression levels of HSP60 in cardiomyocytes.
The following RNAs have other functions:
-
(Choice A) mRNA carries the protein-coding message in DNA within the nucleus to the ribosome in the cytoplasm. This occurs because eukaryotic DNA does not leave the nucleus.
-
(Choice B) Ribosomal RNA (rRNA), along with associated proteins, forms ribosomes.
-
(Choice D) Transfer RNA (tRNA), functions during translation and transfers the correct amino acid to the ribosome; this new amino acid is added to a growing polypeptide chain.
Things to remember:
MicroRNAs are small, noncoding RNA molecules that bind to complementary sequences on target mRNA to degrade the target mRNA or block it from being translated by the ribosome.
Stand Out
with a Top Score in AP Biology
Finish your Unit 6 AP Biology review and continue mastering all units with UWorld. Boost your performance and set yourself apart as a strong candidate for competitive colleges, majors, and scholarships by earning a top score.
Get our all-in-one course today!
- Focused AP Bio Videos
- Print & Digital Study Guide
- 500+ Exam-style Practice Questions
- Customizable Quiz Generator
- Adjustable Study Planner
- Realistic Timed Test Simulation
- Colorful Visual Explanations
- Progress Dashboard
- Smart Flashcards & Digital Notebook
Hear From Our AP Students

UWorld’s service is pretty good and helps provide a lot of explanations on subjects I haven’t been confident on before.

The questions here are the most realistic to the AP tests I've seen so far! I appreciate the ability to customize tests as well.

The best part is that all options are well-explained, telling clearly why they are not the right option.
Frequently Asked Questions (FAQs)
What is AP Biology Unit 6 and why is it important for the exam?
AP Biology Unit 6, titled Gene Expression and Regulation, focuses on how genetic information in DNA is transcribed, translated, and controlled to determine cellular function. It forms the foundation for molecular biology and genetics, appearing frequently in both AP Bio Unit 6 FRQs and MCQs. Topics like transcription, translation, gene regulation, and mutations are heavily tested, making this unit crucial for mastering analytical reasoning and experimental interpretation on the AP Biology exam, including unit 6 AP Biology FRQ.
To prepare effectively:
- Review how DNA and RNA interact to produce proteins.
- Study the regulation mechanisms: operons, epigenetic control, and transcription factors.
- Practice connecting gene expression to phenotype and evolution, a key skill tested under AP Biology gene expression concepts.
These skills help students explain how organisms control gene activity and respond to environmental cues.
UWorld’s AP Biology Unit 6 review simplifies AP Bio gene regulation through annotated visuals and practice sets that mirror College Board expectations. The platform’s adaptive tools guide you through complex molecular processes, helping you learn why certain genes are expressed while others remain silent, essential for scoring high on Unit 6 AP Bio MCQ questions and the overall exam.
What topics are covered in AP Bio Unit 6 Gene Expression and Regulation?
Unit 6 in AP Biology covers the mechanisms that allow cells to use genetic information. Major topics include DNA replication, transcription, RNA processing, translation, and post-translational modifications. It also includes gene regulation in prokaryotes and eukaryotes, mutations, and biotechnology applications like PCR and gene cloning. These topics are central to AP Bio Unit 6 MCQs and FRQs, where students interpret data on gene control, AP Biology gene regulation and protein synthesis.
A structured study order improves comprehension:
- Start with DNA and RNA structure. Understand nucleotide pairing and base differences.
- Move to transcription and translation. Learn how codons specify amino acids.
- Explore gene regulation and epigenetics. Compare operons to histone modification.
- Finish with biotechnology. Focus on genetic engineering and CRISPR concepts frequently reviewed in AP Bio unit 6 test practice materials.
Each concept builds logically on the previous one, forming the foundation for applied reasoning and experimental questions.
How should I study for the AP Biology Unit 6 review?
Studying for AP Biology Unit 6 review requires balancing conceptual understanding with consistent practice. Begin with the College Board Course and Exam Description (CED) to see how gene expression fits into overall exam weighting. This unit frequently appears in data interpretation FRQs and regulation-based MCQs, so active learning is key.
Follow these steps:
- Review all definitions: transcription, translation, and gene regulation.
- Draw process maps for mRNA synthesis and protein production.
- Practice connecting topics like gene expression and phenotype variation.
- Solve AP Bio Unit 6 progress check MCQs to identify weak spots.
Applying knowledge visually and through spaced repetition improves long-term recall and prepares you for question variability.
UWorld’s AP Bio Unit 6 practice test and study tools integrate diagrams, animation-based explanations, and timed quizzes to reinforce both understanding and speed. Combining UWorld with official practice questions ensures conceptual mastery and readiness for the AP Biology unit 6 test.
What’s the best study sequence for AP Bio Unit 6?
A logical study sequence helps you grasp the cause-and-effect relationships within AP Bio Unit 6: Gene Expression and Regulation. Since this unit connects directly to genetics and cell biology, start from molecular foundations and progress toward complex regulation mechanisms.
Recommended sequence:
- DNA Structure and Replication: Understand base pairing and enzyme functions like helicase and DNA polymerase.
- RNA and Transcription: Learn how mRNA, tRNA, and rRNA contribute to protein synthesis.
- Translation: Study codons, anticodons, and ribosomal activity.
- Gene Regulation: Explore operons in prokaryotes and chromatin remodeling in eukaryotes.
- Mutations and Biotechnology: Review the effects of mutations and techniques like CRISPR and PCR.
This order helps solidify both the molecular processes and their biological consequences, preparing you for AP Bio Unit 6 MCQs and FRQs.
What is the most effective AP Bio Unit 6 review plan before the exam?
Creating a structured study timeline helps you master AP Biology Unit 6: Gene Expression and Regulation efficiently. This unit forms the foundation of molecular biology, appearing frequently in AP Bio unit 6 FRQ and unit 6 AP Bio MCQ questions. Breaking your review into 3 stages: 3-months, 1-month, and 2-weeks before the test, ensures you build understanding, retention, and speed. For a complete subject-wide guide, explore our How to Study for AP Biology page.
3-Months Before the Exam:
- Begin with conceptual reading on DNA, RNA, and protein synthesis.
- Watch video lessons on transcription, translation, and AP biology gene expression control.
- Create a topic checklist from the College Board CED and track weak areas weekly.
- End each topic with UWorld’s progress-check sets to reinforce reasoning.
1-Month Before the Exam:
- Focus on advanced topics like mutations, gene regulation, and biotechnology.
- Practice mixed FRQs and MCQs from Units 5 and 6 to connect heredity and expression.
- Summarize terminology and pathways into quick notes or a personalized AP Bio unit 6 cheat sheet.
2-Weeks Before the Exam:
- Simulate timed conditions to build endurance, accuracy, and confidence before the exam.
- Re-attempt progress-check questions on AP Bio gene regulation and challenging MCQs.
- Focus daily on error logs and data-analysis questions similar to UWorld’s AP Biology Unit 6 progress check question formats.
UWorld’s AP Biology Unit 6 review integrates these stages with adaptive analytics, visual explanations, and realistic practice tests. Pairing UWorld with official College Board materials ensures a clear, confident approach to mastering gene expression and regulation before exam day.
What are the main FRQ question types for AP Bio Unit 6?
AP Bio Unit 6 FRQs usually involve experimental analysis, data interpretation, and process explanation. Common themes include gene regulation, mutations, and protein synthesis.
Expect questions that ask you to:
- Interpret graphs showing gene expression changes under different conditions.
- Explain transcription and translation steps using provided sequences.
- Analyze the effects of gene regulation on phenotype or evolutionary advantage.
You might also face synthesis-style questions combining Unit 6 concepts with earlier topics like cellular respiration or evolution. Practicing these will strengthen your analytical reasoning.
How can I use UWorld’s AP Bio Unit 6 progress check questions effectively?
UWorld’s AP Bio Unit 6 progress check questions are designed to help you reflect on your understanding of gene expression and regulation after completing each topic. They act as guided checkpoints, showing how well you’ve grasped the concepts before moving on to the next section.
To use them effectively:
- Treat each set as a learning tool, not a test. Focus on comprehension rather than score.
- Review explanations thoroughly. Each question includes detailed rationales and visuals that clarify why one option is correct and others are not.
- Use performance insights. Identify which question types, like transcription steps or operon control; you find most challenging and revisit related lessons.
- Reattempt questions. Repetition reinforces memory and builds confidence before full-length practice tests.
By approaching progress checks as diagnostic tools, you’ll recognize patterns in how you think and apply biology concepts. This reflection-driven practice deepens understanding and ensures steady progress through AP Biology Unit 6 Gene Expression and Regulation.
UWorld’s adaptive learning system supports this process with real-time analytics and visual explanations, making it easy to track growth, correct misconceptions, and build mastery before formal exam preparation.
How are gene expression and regulation concepts connected in AP Biology Unit 6?
In AP Bio Unit 6, gene expression and regulation are 2 sides of the same process. Expression is the conversion of DNA information into functional proteins, while regulation determines when and how this happens. Understanding this relationship is vital for data-based questions and FRQs.
Consider these links:
- Transcription factors control mRNA production.
- Epigenetic mechanisms like methylation affect gene accessibility.
- Feedback loops regulate enzyme synthesis in metabolic pathways.
Recognizing these patterns helps connect molecular function to observable traits.
How can I retain complex processes or topics in Unit 6?
Retention improves when you link abstract processes to visual and logical cues. For AP Bio Unit 6, focus on mapping each process step-by-step.
Retention strategies:
- Create flowcharts for transcription and translation.
- Use mnemonic devices for operon components (e.g., “PROG”: Promoter, Repressor, Operator, Gene).
- Teach the concept aloud, explaining reinforces memory.
- Review AP Bio Unit 6 notes weekly with self-quizzing.
Consistency and visualization are key. Revisit diagrams until each component’s function becomes second nature.
How do I study gene expression and regulation for the Unit 6 test?
Studying gene expression and regulation effectively involves connecting molecular steps to their biological consequences.
Focus on:
- DNA → RNA → Protein: Know the enzymes, sequences, and purpose of each step.
- Regulation: Study the lac operon, trp operon, and eukaryotic enhancers.
- Mutation impact: Understand how base changes alter proteins.
- Application: Review experimental design and data interpretation questions.
Using mixed question formats (MCQs and FRQs) develops flexibility and reinforces critical thinking.
UWorld’s AP Bio Unit 6 practice test offers timed sets that integrate conceptual and experimental questions. Reviewing explanations deepens your reasoning and helps apply textbook learning to real exam formats, crucial for scoring high in this unit.
What is the best AP Bio Unit 6 practice test format?
The ideal AP Bio Unit 6 practice test includes a blend of MCQs for breadth and FRQs for depth. A balanced simulation mimics the official AP exam’s timing and complexity.
Structure your test as:
- 30–40 MCQs: Mix concept recall and data interpretation.
- 2 FRQs: One experimental and one synthesis-based question.
- 60–70 minutes total: Include a 5-minute review for error marking.
This mirrors the real exam’s mental pacing and ensures exposure to all question types.
When should I begin preparing for the Unit 6 test?
It’s best to start preparing for AP Bio Unit 6: Gene Expression and Regulation as soon as your class begins covering molecular genetics topics. Early engagement allows more time to master concepts like transcription, translation, and gene regulation before your final unit test.
Begin with light review and practice questions during lessons to build a foundation. About two to three weeks before the test, increase intensity by scheduling dedicated study blocks for MCQs, FRQs, and concept maps. If possible, revisit earlier topics such as heredity and cell communication to strengthen cross-unit understanding.
Consistent short sessions are more effective than last-minute cramming. This approach prevents fatigue and helps retain details of processes like mRNA transcription and protein synthesis long term.
What are the common mistakes students make in AP Bio Unit 6?
Common errors in AP Bio Unit 6 often stem from mixing up molecular steps or misunderstanding regulation levels.
Frequent mistakes include:
- Confusing transcription with translation.
- Forgetting RNA processing steps like splicing and poly-A tail addition.
- Misinterpreting operon models (especially lac and trp).
- Overlooking epigenetic modifications in gene regulation.
Awareness of these pitfalls helps improve both accuracy and speed during exams.
UWorld’s error analysis tools highlight trends in your incorrect answers, allowing targeted review. By practicing similar question sets and reviewing rationales, students learn to avoid these recurring mistakes and approach Unit 6 confidently.
Where can I find AP Bio Unit 6 notes, cheat sheets, or study guides?
Reliable AP Bio Unit 6 notes and study guides summarize gene expression and regulation efficiently. Look for sources aligned with the College Board CED, covering transcription, translation, and regulatory mechanisms.
UWorld’s interactive study guide provides condensed yet detailed coverage of every topic, complete with visuals and checkpoints. Supplement it with official College Board unit outlines and Khan Academy summaries for diverse explanations.
For quick review, focus on diagrams and flowcharts that outline molecular pathways. Cheat sheets summarizing operon regulation, codon charts, and mutation types are particularly helpful for exam revision.
Are there downloadable AP Bio Unit 6 study guide PDFs and practice materials?
Yes. You can access downloadable AP Bio Unit 6 study guide PDFs and practice materials that simplify exam review. These typically include summaries of gene expression and regulation, key vocabulary, and flow diagrams illustrating DNA-to-protein conversion.
When selecting downloads, ensure they follow College Board’s content outline. Prioritize resources that include progress check-style questions and answer rationales to apply concepts actively.
UWorld offers printable Unit 6 study guides, digital flashcards, and self-paced quizzes, making it easy to combine written reference with interactive learning. Using UWorld alongside official materials gives you complete coverage, from conceptual review to test simulation, for AP Biology Unit 6 Gene Expression and Regulation.